| |
| Med Image Comput Comput Assist Interv. 2012; 15(Pt 3): 476–84. doi: 10.1007/978-3-642-33454-2_59.Non-local robust detection of DTI white matter differences with small databases Olivier Commowick* and Aymeric Stamm VISAGES : Vision Action et Gestion d'Informations en Santé
, INSERM : U746, Université de Rennes 1 - INRIA, CNRS : UMR6074 / IRISA, Institut des Neurosciences Cliniques de Rennes (INCR), 1, rue Anatole Le Braz 35700 Rennes, FR |
Diffusion imaging, through the study of water diffusion, allows for the characterization of brain white matter, both at the population and individual level. In recent years, it has been employed to detect brain abnormalities in patients suffering from a disease, e.g. from multiple sclerosis (MS). State-of-the-art methods usually utilize a database of matched (age, sex, …) controls, registered onto a template, to test for differences in the patient white matter. Such approaches however suffer from two main drawbacks. First, registration algorithms are prone to local errors, thereby degrading the comparison results. Second, the database needs to be large enough to obtain reliable results. However, in medical imaging, such large databases are hardly available. In this paper, we propose a new method that addresses these two issues. It relies on the search for samples in a local neighborhood of each pixel to increase the size of the database. Then, we propose a new test based on these samples to perform a voxelwise comparison of a patient image with respect to a population of controls. We demonstrate on simulated and real MS patient data how such a framework allows for an improved detection power and a better robustness and reproducibility, even with a small database. MeSH keywords: Brain, pathology, Databases, Factual, Diffusion Tensor Imaging, methods, Humans, Information Storage and Retrieval, methods, Multiple Sclerosis, pathology, Nerve Fibers, Myelinated, pathology, Pattern Recognition, Automated, methods, Radiology Information Systems, Reproducibility of Results, Sensitivity and Specificity |
Diffusion weighted imaging is an MRI modality that provides information about water diffusion within tissues. It has therefore gained much interest for the study of brain white matter architecture. In particular, it may be utilized for the detection of structural differences related to a disease. Reported studies on diseases usually fall within two categories: (i) group comparisons between a population of healthy subjects and a group of patients suffering from the disease and (ii) comparison of one patient to a set of healthy controls. The former aims at characterizing the overall course of a disease while the latter focuses on detecting its early signs and, possibly, its future evolution. Both approaches are of great interest to understand a disease. In this work, we are interested in diffusion imaging for multiple sclerosis (MS). MS is a demyelinating disease, causing both lesions visible on conventional MRI and diffuse damage to the brain white matter architecture that may be visible in diffusion imaging [1]. Having a robust detection of that diffuse damage for a specific patient is crucial as it could help to predict how the disease will evolve in time, and potentially allow to adapt the treatment. Recent works on the comparison of diffusion images have first focused on scalar values extracted from the diffusion tensor, such as mean diffusivity (MD) or fractional anisotropy (FA). For example, Filippi et al. [2] presented a study on manually defined regions of interest demonstrating an MD increase and an FA decrease in specific regions for MS patients brains. However, utilizing only a scalar value may discard a large part of the tensor information and decrease the precision of the comparison. To overcome this problem, several groups have proposed methods to utilize the full tensor either for population comparison (Lepore et al. utilized the Hotelling’s T
2 test on tensors for HIV patients [3], Whitcher et al. [4] the Cramers test on tensors), or for patient to group comparison (Commowick et al. [5]). These works have demonstrated that a test based on the full tensor information yields more precise comparisons. Finally, when high quality data is available (HARDI acquisitions), one may now consider higher order models such as orientation distribution functions (ODFs) to get improved sensitivity in crossing fibers regions where the diffusion tensor performs poorly [6]. Independently of their strengths and weaknesses, comparison methods usually rely on a parametric or permutation statistical test. Such approaches often require large databases either to ensure that the distribution of the test statistic matches the hypothesized one or to make the permutation test data independent. However, in medical imaging studies, databases are usually small due to the difficulty to recruit patients and volunteers, and they may be even smaller when parameters such as age or sex must match between the control database and the patients. In those cases, the chosen statistical test may become erroneous and generate either false positive or false negative detections. In addition, all automatic approaches need a common reference frame that is often constructed from the healthy subjects by means of non linear registration (so called atlas construction [7]). However, such registration methods are not perfect and may be prone to errors due to noise and artifacts. Such errors may further corrupt the comparison performance. To tackle these issues, we propose a new methodology for the robust detection of white matter differences at a patient level. It is based on ideas recently introduced for non-local means denoising [8] and segmentation [9], adapted for a patient-to-group comparison of diffusion models that can be represented as vectors in a vector space (e.g diffusion tensors or ODFs). We present in Section 2 the overall comparison method. We then apply this new method to simulated data and real data of multiple sclerosis patients demonstrating higher accuracy and reproducibility for differences detection over state-of-the-art methods. |
Non-Local Means for the Comparison of Diffusion Data In the following, we assume that a database of M images Im
has been constituted, i.e. all these images have been non linearly registered to a common reference system, and that we are interested in comparing voxel by voxel an image T to the reference database. We propose an algorithm relying on the non-local means framework [10] optimized by Coupé et al. for medical image denoising [8] and segmentation [9]. For each point x of T, we define a patch B(x) (half size h) around it and follow these main steps:
-
For each image Im
, search for patches Bm
(xj
) similar to the patch B(x) of T in a neighborhood N(x) around x
-
Associate a weight wmj
to each patch Bm
(xj
), depending of the similarity between B(x) and Bm
(xj
) (Section 2.1)
-
Keep the center voxel Dmj
of each patch and associate it to its weight wmj
-
Utilize the set of weighted samples to perform the comparison between T and Im, m ∈ {0, …, M} (Section 2.2)
This framework has several advantages: it may help to account for potential registration errors onto the common template for comparison, and it may significantly increase the number of samples to perform the voxelwise comparison even though the database consists of a limited number of images. 2.1. Similarity Weights between Patches The selection of patches is a crucial point as it will define the relative importance of each patch in the final differences detection step. We consider that the model chosen to describe the diffusion of water molecules may be represented as a vector, e.g. tensors in the Log-Euclidean framework [ 11] or ODFs on a spherical harmonics basis [ 6]. Before comparing patches, a preselection is performed for speed reasons and to avoid the degeneracy of the patches weights wmj
:
-
Compute the Log-Euclidean distance between the covariance matrices Σ
B(x) and Σ
B
m
(x
j
): if it exceeds the average distance between any two covariance matrices Σ
B
m
(x) of the database, discard Bm
(xj
), otherwise proceed to the next step;
-
Compute Hotelling’s T
2 statistic [12] to test for mean differences between B(x) and Bm
(xj
) using the pooled covariance matrix: if it exceeds the average statistic computed from any two patches Bm
(x), discard patch Bm
(xj
).
For the remaining patches, we then compute their weights. The weight wmj
between two patches B(x) and Bm
(xj
) is defined as a function of the sum of squared differences between the two patches:
where Δi
= Im
( i + xj
− x) − T( i) are the differences between corresponding voxels of the patches, | B( x)| is the number of voxels in B( x), β a user-defined scale parameter and Σ̂( x) is the local noise covariance around x on T. These weights characterize the similarity between patches and vary between 0 and 1: 1 is reached when the two patches are equal, 0 corresponds to a total disagreement. Since structures with different orientations may occur, the noise covariance Σ̂ is estimated locally. Computing it globally over the whole image could indeed lead to an over-estimation and therefore to biased weights. Coupé et al. [8] proposed a method to estimate such a local noise variance on scalar valued images. Here, we extend it to vector-valued images. Σ̂(x) is estimated from voxels in patch B(x):
-
For each voxel xi
in B(x), consider a small neighborhood of N voxels around it (e.g. the 26 neighbors of xi
). Pseudo-residuals ε
x
i
are computed as:
-
The local image noise covariance is defined from these pseudo-residuals:
2.2. Comparison of Weighted Data Samples We have constituted a list of weighted samples S = { S
1, …, SM
} at each voxel x of T, where Sm
= {( Dm
0, wm
0), …, ( DmJ
, wmJ
)}. We now utilize these samples to confront the patient image to the healthy subjects database. We compute the weighted mean μx
and weighted covariance matrix Σx
at each point x as:
These estimates are very interesting as they take into account the similarity of each patch in the estimation of the mean and covariance. We then test for voxelwise differences by computing the Mahalanobis distance at each point:
where Dx
is the vector value of the patient image at point x (e.g. log-tensor or ODF value). Considering there are enough samples, this squared distance follows a χ
2 distribution with d degrees of freedom, where d is the vector dimension, and a p-value is computed from Z
2 as:
where
 is the cumulative distribution function of a χ
2 distribution with d degrees of freedom. |
Our method has two main parameters: the patch size and the search neighborhood. The smaller the sizes, the closer the method gets to [5]. On the contrary, large sizes tend to increase the number of false negatives. We fixed the parameters on the basis of qualitative results on several patients: patch size of 3 × 3 × 3 (h = 1) and a neighborhood for patch search of 4 voxels in every direction. In addition, we have set β - Eq. (1) - to 1 as is suggested by Coupé et al. [8]. 3.1. Experiments on Simulated Data We first present a quantitative study on simulated images. Starting from a reference diffusion tensor image ( Fig. 1.a), 90 images were simulated by adding Rician noise to the DWI. Then, a patient image was simulated by inserting lesions, i.e. tensors swollen in the two non principal directions. To illustrate the detection power of our method and its robustness to database size, we randomly selected from the 90 images subgroups of 15 to 90 images and used them as the reference database to compare to the simulated patient. Fig. 1 shows the average Dice score results of our method ( M
2) and the one proposed in [ 5] ( M
1). This figure illustrates well the issues arising when using a small database for differences detection. As the sample size decreases, M
1 performs worse, mainly due to a large number of false positives being detected (see Fig. 1.c). These errors mainly stem from the small size of the database that weakens the power of the test. Instead, M
2 obtains much better and more steady scores, which demonstrate its robustness. M
2 performs better as we are able from a small database to increase the number of samples used for the comparison. 3.2. Experiments on Multiple Sclerosis Data We have utilized the LONI ICBM database of healthy control diffusion images 1,. This database is composed of 160 control images: T1-weighted images (isotropic 1 mm
3) and diffusion images acquired on a 3T MRI scanner (b-value of 1000 s/ mm
2, 30 directions with a resolution of 2x2x2 mm
3). This control subject database was compared to a database of 10 MS patient images acquired following a similar protocol with the same parameters. As a first step before processing, the diffusion tensor images are first registered to the T1-w images using a global affine transform [ 13] and a non linear free-form deformation [ 14] with few control points to recover EPI distortions. Then, a DTI atlas is computed from the control subjects DTI using Guimond’s et al. atlas construction method [ 7], combined to a non linear tensor-based registration algorithm. Each DTI patient image is then registered onto the atlas and compared voxel by voxel to the database of controls either with the method proposed in [5] M
1 or the proposed method M
2. We present in Fig. 2 a representative qualitative comparison of the results obtained by the two methods utilizing only a subgroup of 40 images from the controls subjects database. We can notice on this figure that M
1 is affected by the small size of the database and the registration errors, resulting in a large number of false positive detections in Fig. 2.b. On the contrary, adding additional patches as it is done in M
2 leads to many more patches being considered (see Fig. 2.d) and possibly more accurate ones if the registration errors were in the bounds of the local neighborhood. As a consequence, the detection results in Fig. 2.c reveal much less false positives while keeping the detection power on the MS lesions. Finally, we present a quantitative evaluation of the reproducibility of the obtained score maps when the control subjects database changes. To do so, we have, for each patient, repeatedly selected NDb
images out of the 160 images of the database. We have then computed for each of these sub-databases a score map deriving either from M
1 or M
2. To evaluate the variability of the scores, we have chosen to utilize the average of the voxelwise standard deviation of these maps. We present in Table 1 the average over all images of these standard deviation values for NDb
= 20, 40 and 80 images. This table shows that the obtained standard deviations are significantly lower for M
2 (paired t-tests, p-value of 0.001). This indicates a better reproducibility of the results when considering our non-local approach. This confirms the robustness of the proposed method and the interest of utilizing neighboring patches, especially when performing a comparison against a very small database. |
We have presented a new method for the robust detection of differences between a patient diffusion image and a population of control subject diffusion images. It relies on the search for additional patches in a local neighborhood of each voxel utilizing the non-local means framework adapted to diffusion tensor images in the Log-Euclidean space. We have demonstrated both on simulated and real datasets that this allows to detect more accurately differences even if the reference database is small, and to be more robust to potential registration errors. Moreover, it may be applied to any type of diffusion data that can be represented as vector values such as ODFs in a spherical harmonics basis, which should further increase detection performance in regions with crossing fibers. Future works will include an in-depth study of weights definition for oriented structures. The weights may be erroneous in patches where different orientations are present, which could lead to decreased performance. Accounting for these changes in orientations will therefore further improve comparison quality. We will also investigate other approaches to use the selected patches to detect differences. For example, our method could be coupled with a robust comparison algorithm such as the one proposed by Commowick et al. for tensors [15]. Accounting for spatial correlation between the selected patches could also bring further improvements to the comparison. Finally, we will also investigate how to extend our approach to robust population comparison. |
Footnotes |
1.
Rovaris M,
Gass A,
Bammer R,
Hickman SJ,
Ciccarelli O,
Miller D,
Filippi M,
Diffusion MRI in multiple sclerosis
Neurology
65
10
2005
1526
1532
. 2.
Filippi M,
Cercignani M.
Inglese M,
Comi MHG,
Diffusion tensor magnetic resonance imaging in multiple sclerosis
Neurology
56
2001
304
311
. 3.
Lepore N,
Brun CA,
Chou YY,
Chiang MC,
et al.
Generalized tensor-based morphometry of HIV/AIDS using multivariate statistics on deformation tensors
IEEE Transactions on Medical Imaging
27
1
2008
129
141
. 4.
Whitcher B,
Wisco JJ.
Hadjikhani N,
Tuch DS,
Statistical group comparison of diffusion tensors via multivariate hypothesis testing
Magnetic Resonance in Medicine
57
2007
1065
1074
. 5.
Commowick O,
Fillard P.
Clatz O,
Warfield SK,
Metaxas D,
Axel L.
Fichtinger G,
Székely G,
Detection of DTI white matter abnormalities in multiple sclerosis patients
MICCAI 2008
Volume 5241 of LNCS
2008
975
982
. 6.
Goh A,
Lenglet C.
Thompson P,
Vidal R,
A nonparametric riemannian framework for processing high angular resolution diffusion images and its applications to ODF-based morphometry
Neuroimage
56
2011
1181
1201
. 7.
Guimond A,
Meunier J,
Thirion JP,
Average brain models: A convergence study
Computer Vision and Image Understanding
77
2
2000
192
210
. 8.
Coupé P,
Yger P,
Prima S.
Hellier P,
Kervrann C,
Barillot C,
An optimized blockwise non-local means denoising filter for 3D magnetic resonance images
IEEE Transactions on Medical Imaging
27
4
2008
325
441
. 9.
Coupé P,
Manjón JV,
Fonov V.
Pruessner J,
Robles M,
Collins DL,
Patch-based segmentation using expert priors: Application to hippocampus and ventricle segmentation
NeuroImage
54
2
2011
940
954
. 10.
Buades A,
Coll B,
Morel JM,
A review of image denoising, with a new one
Multiscale Model Simul
4
2
2005
490
530
. 11.
Arsigny V,
Fillard P.
Pennec X,
Ayache N,
Log-Euclidean metrics for fast and simple calculus on diffusion tensors
Magnetic Resonance in Medicine
56
2
2006
411
421
. 12.
Anderson T,
An introduction to multivariate statistical analysis
Wiley
2003
. 13.
Ourselin S,
Roche A.
Prima S,
Ayache N,
Delp S,
DiGoia A,
Jaramaz B,
Block matching: A general framework to improve robustness of rigid registration of medical images
MICCAI 2000
Volume 1935 of LNCS
2000
557
566
. 14.
Rueckert D,
Sonoda LL,
Hayes C.
Hill DLG,
Leach MO,
Hawkes DJ,
Nonrigid registration using free-form deformations: Application to breast MR images
IEEE Transactions on Medical Imaging
18
8
1999
712
721
. 15.
Commowick O.
Warfield SK,
A continuous STAPLE for scalar, vector and tensor images: An application to DTI analysis
IEEE Transactions on Medical Imaging
28
6
2009
838
846
. |
 | Fig. 1 Quantitative Detection Power on Simulated Data |
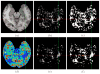 | Fig. 2 Qualitative Comparison on Real MS Patient Images |
 | Table 1 Reproducibility of Comparison Results with Changing Databases |
|








 is the cumulative distribution function of a χ
2 distribution with d degrees of freedom.
is the cumulative distribution function of a χ
2 distribution with d degrees of freedom.