Compacting local protein folds with a Hybrid Protein
Résumé
The "Hybrid Protein Model" (HPM) is a fuzzy model for compacting local protein structures. It learns a non-redundant database encoded in a previously defined structural alphabet composed of 16 protein blocks (PBs). The hybrid protein is composed of a series of distributions of the probability of observing the PBs. The training is an iterative unsupervised process that for every fold to be learnt consists of looking for the most similar pattern present in the hybrid protein and modifying it slightly. Finally each position of the hybrid protein corresponds to a set of similar local structures. Superimposing those local structures yields an average root mean square of 3.14 Å. The significant amino acid characteristics related to the local structures are determined. The use of this model is illustrated by finding the most similar folds between two cytochromes P450.
Fichier principal
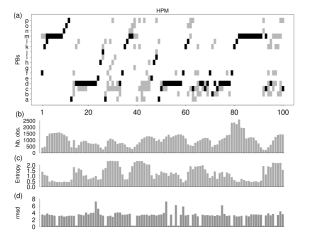 FIGURE_3.PDF (7.43 Ko)
Télécharger le fichier
ARTICLE.pdf (556.53 Ko)
Télécharger le fichier
CAPTIONS.PDF (49 Ko)
Télécharger le fichier
CAPTIONS.pdf (84.16 Ko)
Télécharger le fichier
FIGURE_1.PDF (241.33 Ko)
Télécharger le fichier
FIGURE_1.pdf (275.83 Ko)
Télécharger le fichier
FIGURE_2.PDF (101.02 Ko)
Télécharger le fichier
FIGURE_3.pdf (11.21 Ko)
Télécharger le fichier
FIGURE_4.PDF (22.25 Ko)
Télécharger le fichier
FIGURE_4.pdf (29.31 Ko)
Télécharger le fichier
FIGURE_5.PDF (13.25 Ko)
Télécharger le fichier
FIGURE_5.pdf (17 Ko)
Télécharger le fichier
FIGURE_6.PDF (4.8 Ko)
Télécharger le fichier
FIGURE_6.pdf (8.04 Ko)
Télécharger le fichier
FIGURE_7.PDF (8.09 Ko)
Télécharger le fichier
FIGURE_7.pdf (12.26 Ko)
Télécharger le fichier
TABLE.PDF (19.27 Ko)
Télécharger le fichier
TABLE.pdf (49.78 Ko)
Télécharger le fichier
FIGURE_3.PDF (7.43 Ko)
Télécharger le fichier
ARTICLE.pdf (556.53 Ko)
Télécharger le fichier
CAPTIONS.PDF (49 Ko)
Télécharger le fichier
CAPTIONS.pdf (84.16 Ko)
Télécharger le fichier
FIGURE_1.PDF (241.33 Ko)
Télécharger le fichier
FIGURE_1.pdf (275.83 Ko)
Télécharger le fichier
FIGURE_2.PDF (101.02 Ko)
Télécharger le fichier
FIGURE_3.pdf (11.21 Ko)
Télécharger le fichier
FIGURE_4.PDF (22.25 Ko)
Télécharger le fichier
FIGURE_4.pdf (29.31 Ko)
Télécharger le fichier
FIGURE_5.PDF (13.25 Ko)
Télécharger le fichier
FIGURE_5.pdf (17 Ko)
Télécharger le fichier
FIGURE_6.PDF (4.8 Ko)
Télécharger le fichier
FIGURE_6.pdf (8.04 Ko)
Télécharger le fichier
FIGURE_7.PDF (8.09 Ko)
Télécharger le fichier
FIGURE_7.pdf (12.26 Ko)
Télécharger le fichier
TABLE.PDF (19.27 Ko)
Télécharger le fichier
TABLE.pdf (49.78 Ko)
Télécharger le fichier